Rapid Mycoplasma Detection in Biopharmaceuticals – NAT Method

In recent years, with the rapid development of biopharmaceuticals and the emergence of cell and gene therapies and mRNA vaccines during the pandemic, ensuring the safety and reliability of biopharmaceuticals has become a focal point for regulatory authorities and governments worldwide. Mycoplasma contamination is a common but often challenging type of contamination to eliminate. Regulatory requirements mandate that "absence of mycoplasma contamination must be ensured" for bioprocesses involving cell culture.
Regulatory Authority-required Mycoplasma Detection Points
Raw Materials→Cell Culture Medium→Cell Seed→Cell Culture→Cell Culture Harvest→Purification→Final Product
In the 2020 edition of the Pharmacopoeia of the People's Republic of China Part III, "Preparation and Quality Control of Animal Cell Substrates for Biological Product Production," it is proposed that for production cells, Mycoplasma testing should be conducted on the Master Cell Bank (MCB), Working Cell Bank (WCB), and End of Production Cells (EOPC).
In the "Technical Guidelines for Pharmaceutical Research and Evaluation of Immunocellular Therapy Products (Trial)" it is recommended to conduct Mycoplasma and other safety-related testing on appropriate intermediate samples at key time points or take relevant measures to control them. Mycoplasma testing is also required as a release test for final products.
According to the FDA's "Guidance for Industry: Characterization and Qualification of Cell Substrates and Other Biological Materials Used in the Production of Viral Vaccines for Infectious Disease Indications," Mycoplasma control is required for raw materials, viral seeds, and unprocessed harvest fluids.
Nucleic Acid Testing (NAT) and Traditional Methods
Before the advent of Nucleic Acid Testing (NAT) as a rapid detection method, traditional culture-based methods and indicator cell assays, due to their long detection cycles or sensitivity issues, led to extended production cycles or the need for conditional release of cell substrates based on risk assessment or the search for alternative methods. With the development of cell and gene therapy, the industry's demand for timeliness and sensitivity in pathogen detection has increased. Shorter shelf lives are insufficient to support such long detection cycles, thus NAT methods have emerged as a standout among numerous pathogen detection methods.
Currently, the European Pharmacopoeia (EP) <2.6.7>, Japanese Pharmacopoeia (JP), and United States Pharmacopoeia (USP) <63> have all included NAT methods as pathogen detection methods. However, validation of this method and comparison of its detection sensitivity with traditional methods are required before use. Although the 2020 edition of the Chinese Pharmacopoeia (ChP) does not include NAT as a pathogen detection method, it mentions "other methods approved by the national drug regulatory authorities" can be used.
In May 2022, the Center for Drug Evaluation of the National Medical Products Administration issued the "Technical Guidelines for Pharmaceutical Research and Evaluation of Immune Cell Therapy Products (Trial)" which states that "when the sample size is limited, or rapid release is required, and if pharmacopoeial methods are not suitable, new sterile and pathogen detection methods can be developed for release testing, but these new detection methods should be adequately validated." Therefore, NAT is expected to be increasingly used as a method capable of supporting rapid pathogen release testing by more raw material suppliers and Advanced Therapy Medicinal Product (ATMP) companies.
NAT Method - Requirements for Method Validation
The validation of NAT methods is detailed in both the European Pharmacopoeia <2.6.7> and the Japanese Pharmacopoeia, with their content being essentially consistent. The National Institutes for Food and Drug Control (NIFDC) has also published an article titled "Considerations on Nucleic Acid Detection Methods and Methodological Validation for the Detection of Pathogens," aiming to provide guidance for developers and users of nucleic acid amplification methods for pathogen detection in China. Specificity, detection limit, and robustness validation are required for NAT methods for pathogen detection. If commercial assay kits are used for pathogen detection, appropriate method applicability validation can be conducted based on the supplier's comprehensive assay kit performance validation report combined with the laboratory environment and sample types.
Specificity
Specificity refers to the ability of an analytical method to accurately measure the target analyte in the presence of other components such as impurities, degradation products, excipients, etc. In the European Pharmacopoeia (EP), it is mentioned that during validation, attention should be paid to cross-reactivity between closely related bacterial species, such as Gram-positive bacteria including species from Clostridium, Lactobacillus, and Streptococcus genera. Additionally, common types of human DNA contamination in host cells and laboratory operational environments should also be considered.
Detection Limit
Detection limit refers to the lowest amount of the analyte in the sample that can be detected. The detection limit serves as a criterion for limit tests and qualitative identification and does not need to be quantified as an exact value. In the European Pharmacopoeia (EP), it is required that for each pathogen and each dilution concentration, there should be 24 detection data points. This can be achieved by conducting three independent 10-fold serial dilutions on different days, with 8 repetitions for each dilution gradient, or by conducting four independent 10-fold serial dilutions on different days, with 6 repetitions for each dilution gradient. A detection positivity rate of over 95% is required to establish the detection limit.
For pathogen types requiring confirmation of the detection limit, assay kit manufacturers should cover as many pathogen types as possible as required by regulatory pharmacopoeias and explore the detection limit for each pathogen. For biopharmaceutical companies, it is typically necessary to select based on the actual sources of raw materials being used. Additionally, consideration should be given to human-derived pathogen types.
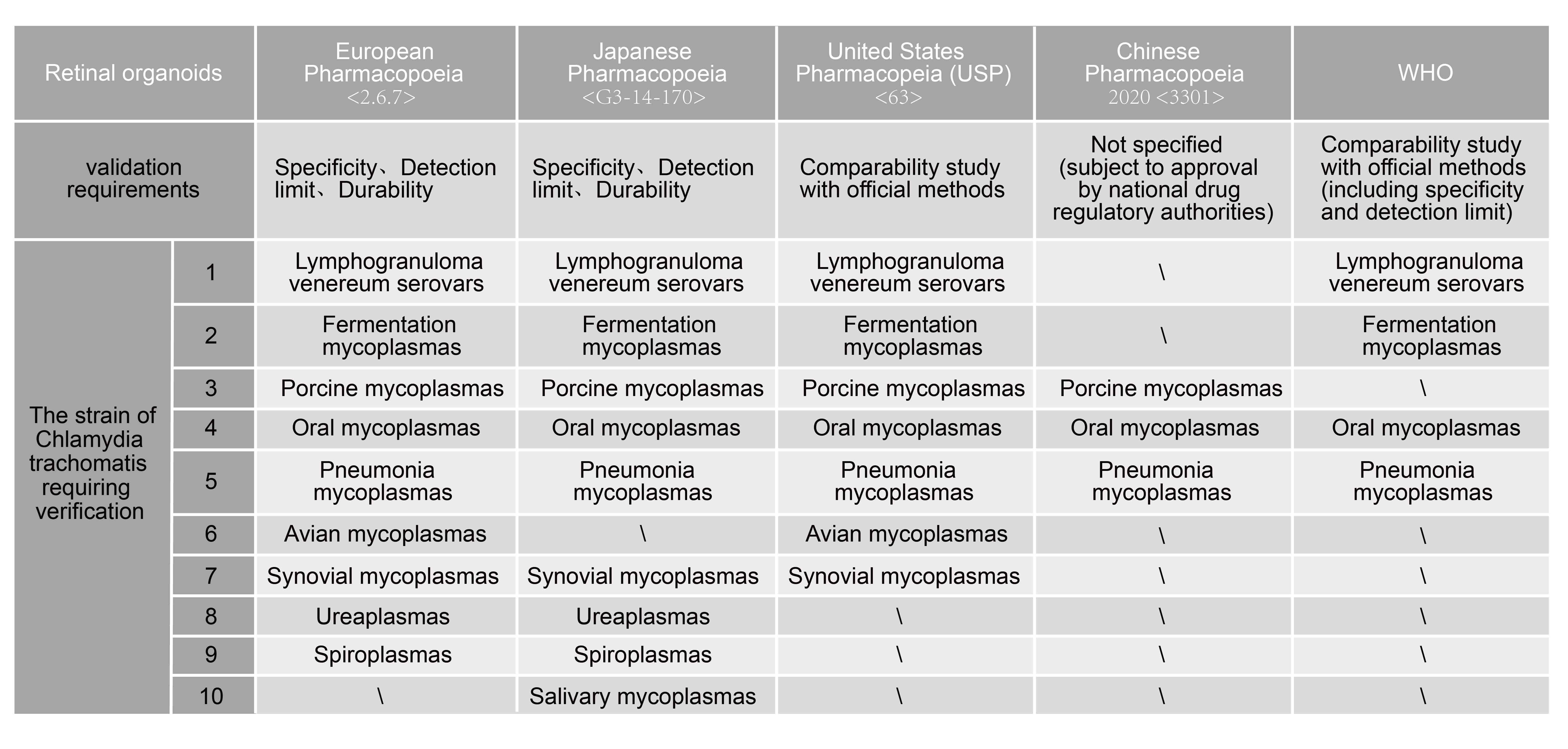
If poultry cells or materials are used or encountered during the production process, validation of the detection limit for Mycoplasma synoviae should be conducted.
If insects or plant materials are used or encountered during the production process, validation of the detection limit for Spiroplasma should be conducted.
Porcine nasal mycoplasma is highly prevalent in mycoplasma-contaminated samples and has received attention from the China National Institutes for Food and Drug Control.
In contrast to the European Pharmacopoeia, the Japanese Pharmacopoeia does not mention Mycoplasma gallisepticum for validation of the detection limit. Instead, it includes Mycoplasma orale in addition to the mentioned criteria.
Robustness
Robustness refers to the degree to which the results of a measurement remain unaffected by small variations in the measurement conditions, providing a basis for the established method's use in routine testing. For the evaluation of robustness, attention needs to be given to the tolerance of the pathogen detection method to changes in the concentrations of MgCl2, primers, and dNTPs in the detection reagents, variations in nucleic acid extraction kits or extraction steps, and the tolerance to the use of different nucleic acid amplification instruments.
Comparability Study
If NAT is to be used as a replacement for pharmacopoeial methods, it needs to be confirmed through comparison that NAT can substitute for culture-based methods or indicator cell culture methods. Typically, this involves considering the method's detection limit as well as specificity (such as the range of covered pathogens and potential false positives). For the comparison of detection limits, it must meet the criteria of "a detection limit of 10 CFU/mL can replace culture-based methods; a detection limit of 100 CFU/mL can replace indicator cell culture methods."
There are two optional approaches for conducting comparative studies:
1. Conduct synchronous experiments using the same validated pathogen strains for both NAT and pharmacopoeial methods to confirm the detection limit.
2. Compare the validation results of NAT with those of the pharmacopoeial methods, but careful documentation of the confirmation documents for the pathogen standards used in both methods is required.
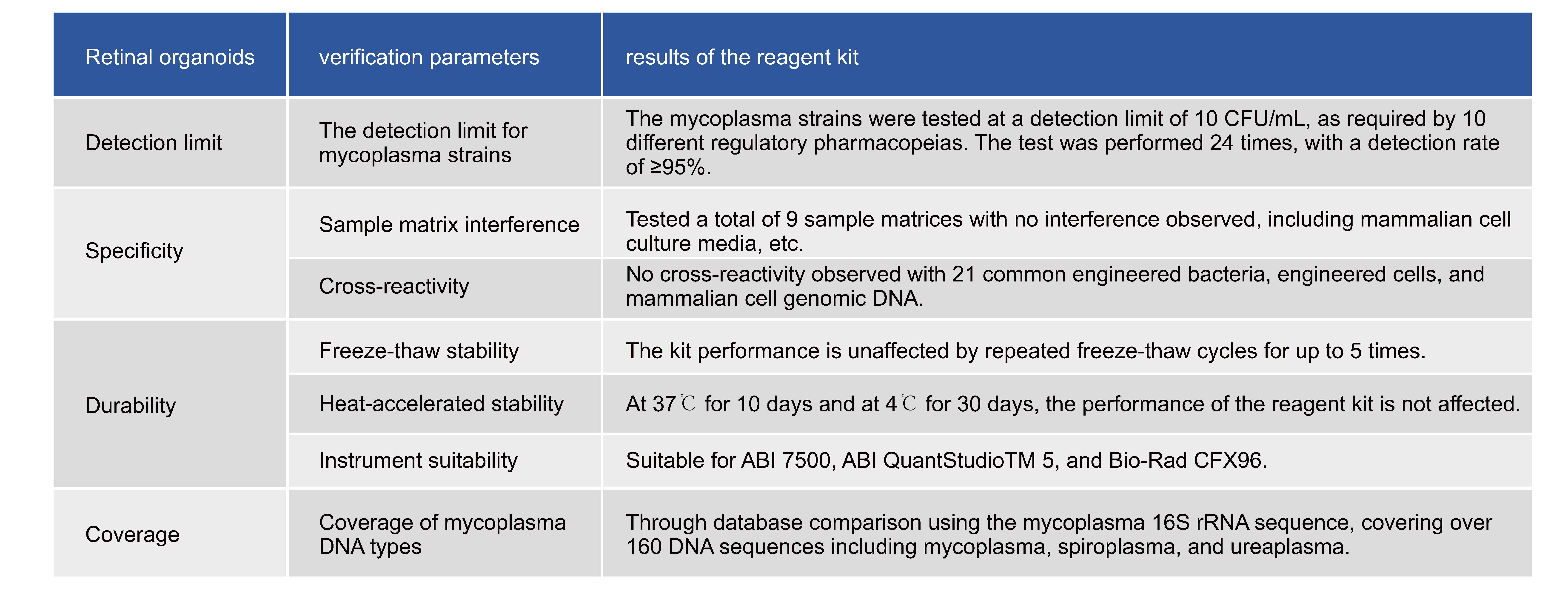
Verification Content of DfCell Mycoplasma Detection
Related products
The Arcegen Bioscience DfCell Mycoplasma qPCR Detection Kit (Probe Method) is a rapid qualitative detection solution based on Nucleic Acid Amplification Techniques (NAT). It is designed for detecting potential mycoplasma contamination in raw materials, cell banks, viral seeds, viral or cell harvest fluids, and therapeutic cells. This kit utilizes quantitative PCR technology and Taqman fluorescent probes to qualitatively detect mycoplasma DNA in the test samples, covering over 160 different mycoplasma DNA sequences. It has been rigorously validated in accordance with the guidelines and requirements of EP 2.6.7 and JP G3 for mycoplasma detection, demonstrating high sensitivity, excellent specificity, and safety.
It can be used in conjunction with the Magnetic Bead-based residual DNA sample pretreatment kit for manual extraction or automated nucleic acid extraction using the Auto-Pure 32A fully automated nucleic acid extraction instrument. After pretreatment to remove interfering impurities and obtain purified mycoplasma DNA, the fluorescent signal from the probes is collected via qPCR, allowing for determination of the detection results.
|
Product Name |
Catalog Number |
Specifications |
|
C230106E/S |
25 T/100 T |
